Acute Kidney Injury and Antimicrobial Stewardship
Acute Kidney Injury and Antimicrobial Stewardship
An estimated 1 in 5 hospital admissions in the UK is associated with acute kidney injury1, providing a clear illustration of the need for novel, rapid detection methods. Our latest whitepaper looks at this common condition and the links between Acute Kidney Injury and Antimicrobial Stewardship. For more details on the things discussed in this article, you can download the full whitepaper below.
Acute Kidney Injury
Acute Kidney Injury is defined as a sudden loss of kidney function. This causes a disruption in the kidneys’ ability to filter waste out of your blood resulting in an accumulation of waste products as well as other imbalances.
The loss of kidney function is the result of a sudden reduction in glomerular filtration rate (GFR), the process through which waste is extracted from the blood and is often reversible2.
Aetiology of Acute Kidney Injury
The differential pressure existing between the glomerulus and Bowman’s is the driving mechanism for glomerular filtration2. This pressure contrast is influenced by the combined resistances of the afferent (leading to the glomerulus) and efferent (leading away from the glomerulus) vascular pathways in the kidney. Under normal kidney function, these resistances are in equilibrium, facilitating the proper functioning of the GFR. For example, an increase in efferent resistance restricts the blood flow out of the kidney, elevating pressure inside the kidney and reducing GFR, and vice versa2. However, in AKI, the decline in renal blood flow and GFR has a pathological origin. The pathophysiology of AKI can be classified as prerenal, intrinsic renal, or postrenal.
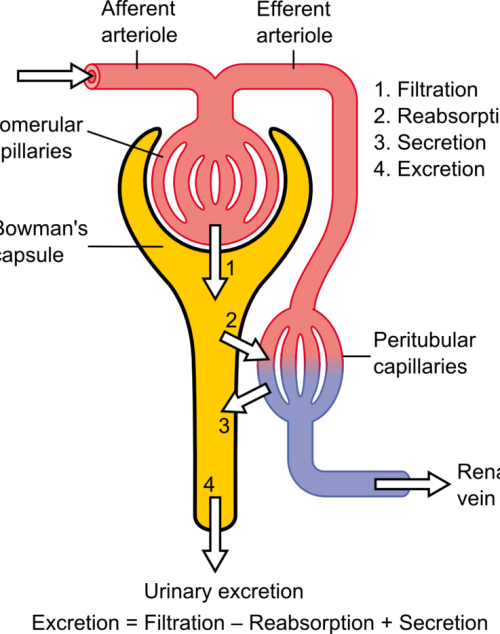
Pre-renal Acute Kidney Injury
Pre-renal AKI is caused by reduced afferent blood flow or, in other words, increased afferent resistance. While tubular and glomerular functions generally remain intact, pre-renal AKI may be caused by systemic hypoperfusion (decreased blood flow) or selective hypoperfusion to the kidney, caused by renal artery stenosis or aortic dissection3.
Intrinsic Renal Acute Kidney Injury
Renal AKI describes the conditions which affect the glomerulus or tubule, for example, acute tubular necrosis and acute interstitial nephritis. This collection of conditions is associated with vasoconstrictor expression in renal afferent pathways2.
Post-renal Acute Kidney Injury
Post-renal AKI usually results from an obstruction in the filtration system. Causes of obstruction include kidney stones, tumours, or blood clots, commonly in the bladder outlet. Obstruction affecting one side might not invariably lead to acute kidney injury, especially when the impediment develops slowly, such as with tumour growth. This is because the unaffected kidney might be able to adjust and make up for the compromised functionality3
Symptoms & Treatment
AKI often manifests with decreased urine output as its primary symptom. However, additional symptoms, when present, can encompass fatigue, nausea, vomiting, or confusion4. To achieve an accurate diagnosis, a comprehensive review of the patient’s medical history and a physical examination are essential to identify the underlying cause of the condition.
The treatment and management of AKI are contingent upon the root cause of the condition. In milder cases, measures are implemented to maintain appropriate levels of fluid, electrolytes, and blood pressure. Nutritional support may also be administered when necessary. In the most severe instances of AKI, dialysis may be warranted to compensate for the diminished kidney function5.
Creatinine serves as a valuable diagnostic tool for evaluating renal conditions, including kidney health, GFR, and muscular dystrophy. However, abnormal serum creatinine (SCr) levels only become evident when a significant portion of the renal mass is compromised. The kidneys possess an impressive capacity to adapt to reduced function, which means that a considerable loss of function or GFR is necessary to influence SCr levels. This poses a challenge when it comes to early detection of AKI6.
Novel biomarkers, KIM-1, NGAL, Clusterin, and Cystatin C, are associated with AKI2 and can be analysed through molecular testing. These new methods can provide a fast and accurate assessment of an individual’s kidney health, at a much earlier stage than SCr quantification2.
Antimicrobial Stewardship
Antimicrobial Stewardship (AMS) programs are specifically crafted to enhance the efficiency of antimicrobial utilization, curtail the emergence of Antimicrobiasl Resistance (AMR), and enhance patient outcomes7. These programs encompass a variety of approaches, such as educational initiatives, training, the establishment of guidelines and protocols, ongoing monitoring and feedback regarding antimicrobial usage, and the management of antimicrobial formularies. Through the promotion of prudent antibiotic utilization, AMS programs contribute to the safeguarding of the efficacy of currently available antimicrobial agents and the deceleration of AMR development7.
Antibiotics and Acute Kidney Injury
Various antibiotics are associated with the progression of AKI due to their nephrotoxicity which can cause severe damage to the kidneys. These antibiotics include polymyxins, aminoglycosides and the commonly used, vancomycin8.

Randox Renal Injury Detection
Using the patented Biochip Technology, the Randox Acute Kidney Injury (AKI) array, available on the Evidence Investigator, simultaneously tests for four novel biomarkers (KIM-1, NGAL, Clusterin, Cystatin C) delivering an early diagnosis and monitoring of treatment efficacy. Multiplex testing better captures reduced renal function, as each biomarker reflects different mechanisms that result in similar injury outputs, allowing for a more accurate picture of the underlying cause of AKI. Along with being able to identify AKI at a much earlier stage, this array provides an accurate and sensitive solution for the diagnosis and monitoring of AKI.
If you’d like some more information on the Randox Acute Kidney Injury Array or would like to add this technology to your laboratory, take a look at our website at https://www.randox.com/acute-kidney-injury/ or get in touch today at marketing@randox.com.

References
- NICE. How common is it? Acute Kidney Injury . Published July 2023. Accessed October 2, 2023. https://cks.nice.org.uk/topics/acute-kidney-injury/background-information/prevalence/
- Adiyanti SS, Loho T. Acute Kidney Injury (AKI) Biomarker.; 2012.
- Manzoor H, Bhatt H. Prerenal Kidney Failure.; 2023.
- NHS. Acute Kidney Injury. NHS. Published 2023. Accessed July 31, 2023. https://www.nhs.uk/conditions/acute-kidney-injury/
- Goyal A, Daneshpajouhnejad P, Hashmi M, Bashir K. Acute Kidney Injury . In: StatPearls [Internet]. StatPearls Publishing ; 2023.
- Rule AD, Lieske JC. The estimated glomerular filtration rate as a test for chronic kidney disease: Problems and solutions. Cleve Clin J Med. 2011;78(3):186-188. doi:10.3949/ccjm.78a.11004
- Baur D, Gladstone BP, Burkert F, et al. Effect of antibiotic stewardship on the incidence of infection and colonisation with antibiotic-resistant bacteria and Clostridium difficile infection: a systematic review and meta-analysis. Lancet Infect Dis. 2017;17(9):990-1001. doi:10.1016/S1473-3099(17)30325-0
- Clifford KM, Selby AR, Reveles KR, et al. The Risk and Clinical Implications of Antibiotic-Associated Acute Kidney Injury: A Review of the Clinical Data for Agents with Signals from the Food and Drug Administration’s Adverse Event Reporting System (FAERS) Database. Antibiotics. 2022;11(10):1367. doi:10.3390/antibiotics11101367
Differentiating Viral from Bacterial Infections
Estimates claim that over 1.2 million people died in 2019 as a direct result of an antibiotic-resistant bacterial infection. Statistics show that up to 4.95 million deaths in the same year were associated with antimicrobial resistance (AMR)1. The overuse and misuse of antibiotics is considered to be the largest contributing factor to the rise of AMR. Antibiotics are effective at treating a wide range of bacterial infections, however, when used to treat viral infections, they have little to no effect. Even still, many physicians continue to prescribe so-called empirical antibiotics as an all-encompassing treatment strategy. In their defence, differentiating viral from bacterial infections can be troublesome. Traditional testing takes the form of paired serology, which requires patients to visit a healthcare facility twice during a 2–4-week period. Many of these infections have distressing symptoms, making this an unreasonable time-to-diagnosis period. Novel molecular techniques can reduce the time to result in the determination of many infections. However, some of these methods are associated with high false positive rates and low specificity resulting in further misuse of antibiotics.
Mxyovirus resistance protein A (MxA) is a biomarker associated with viral infections. It displays antiviral activity against positive, double-stranded RNA viruses and some DNA viruses2. In a study from earlier this year, MxA was used to differentiate viral from bacterial infections in a cohort of 61 adults with an AUROC of 0.9 and a sensitivity and specificity of 92.3% and 84.6% respectively3. An additional study, known as the TREND study, found that a cut-off of 430μg/L could effectively differentiate bacterial and viral infections with an AUROC of 0.9, a sensitivity of 92% and a specificity of 100%4.
C-reactive protein (CRP) is a non-specific acute phase protein which is associated with bacterial infection. However, CRP levels have also been shown to be elevated in response to various viral infections such as Influenza virus, malaria5 and SARS-COV-26, limiting its utility in differentiating the aetiology of an infection.
Using both biomarkers in combination can help physicians determine the true aetiology of infection with high specificity, supporting antimicrobial stewardship and reducing the harmful use of these drugs. Available on the VeraSTAT, Randox provides tests for MxA and CRP, which together provide a fast and accurate method of detection and differentiation of bacterial and viral infections from a small sample.
Alternatively, don’t hesitate to browse our range on our website or get in touch with one of our team at marketing@randox.com who will be happy to help with any query you have!
References
- Murray CJL, Ikuta KS, Sharara F, et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. The Lancet. 2022;399(10325):629-655. doi:10.1016/S0140-6736(21)02724-0
- Liao S, Gao S. MxA: a broadly acting effector of interferon-induced human innate immunity. Visualized Cancer Medicine. 2022;3:2. doi:10.1051/vcm/2022002
- Metz M, Gualdoni GA, Winkler HM, et al. MxA for differentiating viral and bacterial infections in adults: a prospective, exploratory study. Infection. Published online February 3, 2023. doi:10.1007/s15010-023-01986-0
- Rhedin S, Eklundh A, Ryd-Rinder M, et al. Myxovirus resistance protein A for discriminating between viral and bacterial lower respiratory tract infections in children – The TREND study. Clinical Microbiology and Infection. 2022;28(9):1251-1257. doi:10.1016/j.cmi.2022.05.008
- Joseph P, Godofsky E. Outpatient Antibiotic Stewardship: A Growing Frontier—Combining Myxovirus Resistance Protein A With Other Biomarkers to Improve Antibiotic Use. Open Forum Infect Dis. 2018;5(2). doi:10.1093/ofid/ofy024
- Paranga TG, Pavel-Tanasa M, Constantinescu D, et al. Comparison of C-reactive protein with distinct hyperinflammatory biomarkers in association with COVID-19 severity, mortality and SARS-CoV-2 variants. Front Immunol. 2023;14. doi:10.3389/fimmu.2023.1213246
RX Imola: Inflammatory Biomarkers in COVID-19
Over the course of human history, few events have had such a dramatic impact as the COVID-19 pandemic. According to the World Health Organization (WHO), as of 12th July 2023, the SARS-CoV-2 virus has claimed almost 7 million lives and figures continue to rise1. While many who become infected are only subject to mild symptoms, those who develop a more severe form of the infection are encumbered with a debilitating flu-like condition, often requiring days, if not weeks of bed rest. In a paper from June 20232, the Rx Imola was used to study C reactive protein concentrations, along with other biomarkers, in mild and severe COVID-19 patients in order to develop novel risk stratification methods for this potentially life-threatening viral infection.
The impact on healthcare services around the world cannot be understated. In developed countries, access to services for both COVID-related and other conditions took a catastrophic hit. In low-to-middle-income countries, the impact has been even more distressing, all but eliminating basic medical care in favour of combating COVID-19, partly due to inferior resources and facilities3.
In times of medical emergency, it is crucial to have an efficient and effective means of stratifying the risk to patients and a process for suitably categorising those into the least and most at risk of severe complications or death. Due to the rate at which COVID-19 spread, unfortunately, the world lacked these mechanisms for SARS-CoV-2, resulting in mass hospital overpopulation, cancelled appointments for other life-threatening conditions and ultimately the staggering mortality statistics we’ve been bombarded with since January 2022. This prompted an unprecedented surge in medical research and major advances in testing capabilities, giving us new methods of detecting SARS-CoV-2 and determining the risk posed to individuals.
One such investigation, by Paranga et al., (2023) studied a total of 13 biomarkers to determine which could accurately differentiate mild, moderate, and severe cases and identify biomarkers which were good predictors of fatality2. C reactive protein (CRP) was the best-described biomarker relating to COVID-19 throughout the pandemic. This paper compares it to 12 other biomarkers including suPAR, sTREM-1, ferritin, MCP-1 and Lactate dehydrogenase. Of these, it was discovered that CRP was clearly the most effective biomarker for differentiating mild from severe cases, with concentrations in those with severe infection being, on average, 45% higher than in those with mild symptoms2. Additionally, the authors discovered that CRP levels were not significantly affected by age, a factor known to affect the inflammation and immune responses, providing a powerful and inclusive risk stratification tool. Some of the additional conclusions drawn from this paper can be seen below2:
- Lactate dehydrogenase, sTREM-1 and HGF were good predictors of mortality in COVID-19.
- suPAR was identified as a crucial molecule in characterising Delta variant infection and mortality.
- The initial values of inflammatory biomarkers were good to excellent predictors of disease severity in COVID-19 patients.
- Disease severity and mortality are associated with a higher rate of comorbidities including thrombocytopenia and other blood diseases, circulatory and respiratory system diseases and liver diseases such as cirrhosis.
So, what is CRP and how does it become elevated in response to a SARS-CoV-2 infection? CRP is a non-specific, acute-phase protein, meaning its concentration is altered in response to inflammation4. The acute respiratory distress syndrome induced by SARS-CoV-2 is, in part, a result of the hyperinflammation caused by the virus2. CRP is a well-characterised inflammatory biomarker and is therefore well-suited for identification and risk stratification in an emerging disease.
This investigation2 utilised the RX Imola, a rapid, comprehensive clinical chemistry platform, to quantify CRP. With the RX Imola, laboratories can gain access to the world’s largest clinical chemistry test menu covering routine chemistries as well as specific proteins, lipids, and more providing a cost-effective and user-friendly platform. With 60 cooled reagent positions and a sample carousel with 20 cooled positions for controls and calibrators, the RX Imola is an ideal solution for small to medium-throughput laboratories seeking an innovative and reliable clinical chemistry system. Randox also supplies suitable, high-quality reagents, and through Acusera, state-of-the-art controls and calibrators, completing the clinical chemistry portfolio.
References
1. World Health Organisation. WHO Coronavirus (COVID-19) Dashboard. https://covid19.who.int/.
2. Paranga TG, Pavel-Tanasa M, Constantinescu D, et al. Comparison of C-reactive protein with distinct hyperinflammatory biomarkers in association with COVID-19 severity, mortality and SARS-CoV-2 variants. Front Immunol. 2023;14. doi:10.3389/fimmu.2023.1213246
3. Jain P. Impact of COVID-19 Pandemic on Global Healthcare Systems and the role of a new era of global collaborations. Sushruta Journal of Health Policy & Opinion. 2021;14(3):1-5. doi:10.38192/14.3.2
4. Nehring S. C Reactive Protein . https://www.statpearls.com/articlelibrary/viewarticle/18744/.
Randox Clinical Laboratory Services
Randox Clinical Laboratory Services
Randox Clinical Laboratory Services (RCLS) specialises in offering esoteric biomarker testing utilising the expertise and knowledge Randox has built over 40 years producing high quality IVD diagnostics. RCLS strives to provide a clinical laboratory service to meet the time sensitive, bespoke requirements of research and clinical trial projects globally.
By utilising Randox Clinical Laboratory Services you can save your laboratory time and money. Our multiplex panels offer a cost effective, labour saving and time reducing solution for your sample testing. As a result, we provide your laboratory with the best solution for your sample testing.
Clinical laboratories traditionally perform diagnostic testing from a variety of patient samples (whole blood, serum, plasma, saliva, sputum, CSF, swab collections) to determine their health status.
In addition to Diagnostics, Randox Laboratories also perform Biomarker analysis from selected patient cohorts in support of pharma research, aiming at discovering a biological signal related to an NDE’s activity (targeted Biomarker approach) or a novel druggable target (proteomic analysis). Randox Biosciences offer multiple technical solutions to address the above, mostly based on the Randox proprietary Biochip Array Technology.
Why RCLS?


Sample and Data Management
- LIMS system (Labware) utilised and connected throughout the lab
- Customer and project specific folders on an internal secure server
- Ability to transfer data securely through secure email/drop box
Capabilities
Randox Biosciences provide the building blocks of diagnostic development which includes antibodies, corresponding conjugates and proteins. Our immunoassay product offering provides diagnostic solutions covering a variety of disease areas including thyroid, reproduction, cardiovascular and renal.
Biochip Technology from Randox is an innovative assay technology which utilises multiplex testing methodology in a rapid, accurate and easy-to-use format, catering both molecular and immunoassay testing. The technology works by combining a panel of related assays in a single biochip with a single set of reagents, controls and calibrators. Only one single undivided sample is used. The results are read by a CCD-camera and custom image-processing software.
Custom Assay Development
Serving the diagnostic, pharmaceutical, contract research and biotech industries, Randox Biosciences dedicated custom unit develops and manufactures custom assay solutions. This also includes full customised quality control sera and calibrators. A selection of over 320 pre-qualified biomarkers is available for a custom panel design. However, any specific biomarker of interest will be considered for integration into an existing or desired panel, for technical feasibility (concentration range compatibility, potential interferences/cross-reactivities). The number of the required novel biomarkers will help make the feasibility study as a pertinent and complete as possible.
Companion Diagnostics (CDx) are part of the services that Randox Biosciences supply to pharmaceutical companies. With a long-recognized expertise in the development of esoteric assays, and more than 500 IVD products available globally, Randox is a partner of choice to support your CDx needs.
Full Assay Menu
| Acute Kidney Injury | |||
| Clusterin | Cystatin C | Kidney Injury Molecule-I | Lipocalin |
| Adhesion Molecules | |||
| E-Selectin | Intercellular Adhesion Molecule-I | L-selectin | P-selectin |
| Vascular Cell Adhesion Molecule-I | |||
| Anaemia | |||
| Ferritin | |||
| Alzheimers Risk | |||
| ApoE4 | Pan ApoE | ||
| Cerebral | |||
| D-Dimer | Neuron Specific Enolase | Neutrophil Gelatinase-Associated Lipocalin | Soluble Tumour Necrosis Factor Receptor I |
| Chronic Kidney Disease | |||
| C3a Des Arg | C-Reactive Protein | Cystatin C | D-Dimer |
| Epidermal Growth Factor | Fatty Acid-Binding Protein I | Interlukin-8 | Macrophage Inflammatory Protein-1α |
| Neutrophil Gelatinase-Associated Lipocalin | Soluble Tumour Necrosis Factor Receptor I | Soluble Tumour Necrosis Factor Receptor II | |
| Covid-19 | |||
| SARS-CoV-2 Antigen | Receptor Binding Domain | Nucleocapsid | |
| Cytokines | |||
| Granulocyte-Macrophage Colony Stimulating Factor | Interleukin-1α | Interleukin-1ϐ | Interleukin - 2 |
| Interleukin - 3 | Interleukin - 4 | Interleukin - 5 | Interleukin - 6 |
| Interleukin - 7 | Interleukin - 8 | Interleukin - 10 | Interleukin - 12p70 |
| Interleukin - 13 | Interleukin - 15 | Interleukin - 23 | Interferon Gamma |
| Human EGF | Monocyte Chemotactic Protein | Macrophage Inflammatory Protein-1α | Matrix Metalloproteinase-9 |
| Soluble IL-2 Receptor α | Soluble IL-6 Receptor | Soluble Tumour Necrosis Factor Receptor I | Soluble Tumour Necrosis Factor Receptor II |
| Tumour Necrosis Factor Alpha | Vascular Endothelial Growth Factor |
| Epidermal Growth Factor (EGF) | |||
| Amphiregulin (AREG) | Herapin-Binding EGF-like Growth Factor (HBEGF) | Transforming Growth Factor - Alpha (TGF-α) | |
| Gastrointestinal | |||
| Gastrin 17 | Helicobacter Pylori | Pepsinogen I | Pepsinogen II |
| Immunodeficiency | |||
| Interleukin - 17A | Interleukin - 17F | Interleukin - 22 | Interleukin - 17 |
| Metabolic | |||
| Ferritin | Interleukin - 6 | Insulin | Adiponectin |
| C-Reactive Protein | Cystatin C | Leptin | Parathyroid Hormone |
| Plasminogen Activator Inhibitor-I | PTH | Resistin | Tumour Necrosis Factor α |
| Pancreatic Cancer | |||
| Cancer Antigen 19-9 - CA19-9 | Carcinoembryonic Antigen | Alpha-I-Acid Glycoprotein | |
| Stroke | |||
| Brain-Derived Neurotrophic Factor - BDNF | D-Dimer | Glial Fibrillary Acidic Protein - GFAP | Glutathione S - Transferase Pi - GSTPi |
| Heart-Type Fatty Acid-Binding Protein - FABP3 | Interleukin-6 - IL-6 | Nucleoside Diphosphate Kinase - NDKA | Neuron Specific Enolase - NSE |
| Parkinson Protein 7 - PARK-7 | Soluble Tumour Necrosis Factor Receptor I - sTNFRI |
| Thyroid | |||
| Anti-Thyroglobulin (TgAb) | Anti-Thyroid Peroxidase Antibody (TPOAb) | Thyroxine Binding Globulin (TBG) | Total Thyroxine (TT4) |
| Total Tri-Iodothyronine (TT3) | |||
| Tissue Damage | |||
| Adipose Fatty Acid Binding Protein - FABP4 | Brain Fatty Acid Binding Protein - FABP7 | Epidermal Fatty Acid Binding Protein - FABP5 | Heart-Type Fatty Acid-Binding Protein - FABP3 |
| Ileal Fatty Acid Binding Protein - FABP6 | Liver Fatty Acid Binding Protein-I - FABPI | Testis Fatty Acid Binding Protein - FABP9 |
| CARDIAC RISK PREDICTION | |||
| Target | SNP | Target | SNP |
| MIA3* | rs17465637 | LPA* | rs3798220 |
| 9p21 | rs10757274 | LPA* | rs10455872 |
| DAB2IP | rs7025486 | MRAS* | rs9818870 |
| CXCL12 | rs1746048 | LPL* | rs328 |
| ACE | rs4341 | LPL | rs1801177 |
| NOS3 | rs1799983 | SORT1*+ | rs646776+ |
| APOA5 | rs662799 | PCSK9 | rs11591147 |
| SMAD3 | rs17228212 | APOE* | rs429358 |
| APOB* | rs1042031 | APOE* | rs7412 |
| CETP | rs708272 | ||
| CHRONIC LUNG DISEASE (CLD) ARRAY | |||
| Viral | |||
| Adenovirus | Metapneumovirus | Respiratory syncytial virus A | Respiratory syncytial virus B |
| Rhinovirus A/B/C | Influenza virus A | Influenza virus B | |
| Bacterial | |||
| Achromobacter xylosoxidans | Bordetella pertussis | Burkholderia cepacia complex (21spp) | Burkholderia cenocepacia |
| Chlamydia pneumoniae | Haemophilus influenza | Moraxella catarrhalis | Mycoplasma pneumoniae |
| Non-tuberculous mycobacterium (15 spp) | Mycobacterium avium complex (4 spp) | Pandoraea species (5 spp) | Prevotella species (16 spp) |
| Pseudomonas aeruginosa | Staphylococcus aureus | Stenotrophomonas maltophilia | Streptococcus pneumoniae (21 spp) |
| Streptococcus species (19 spp) | Veillonella species (3 spp) | ||
| Fungal | |||
| Aspergillus fumigatus | Candida albicans | Exophialia dermatitidis | Scedosporium species (7 spp) |
| Antibiotic Resistance Markers | |||
| mecA (incl MRSA) |
| FAMILIAL HYPERCHOLESTEROLEMIA | |||
| LDLR 38 mutations | APOB 1 mutation | PCSK9 1 mutation | |
| APOB | PCSK9 | ||
| Mutation | Protein | Mutation | Protein |
| c.10580G>A | p.(Arg3527Gln) | c.1120G>T | p.(Asp374Tyr) |
| LDR | |||
| Mutation | Protein | Mutation | Protein |
| c.2292delA | p.(Ile764Metfs*2) | c.1187-10G>A | p.(=) |
| c.1444G>A | p.(Asp482Asn) | c.1048C>T | p.(Arg350*) |
| c.551G>A | p.(Cys184Tyr) | c.118delA | p.(Ile40Serfs*166) |
| c.1845+11C>G | p.(=) | c.1168A>T | p.(Lys390*) |
| c.693C>A | p.(Cys231*) | c.232C>T | p.(Arg78Cys) |
| c.933delA | p.(Glu312Serfs*58) | c.1587-1G>A | p.(=) |
| c.301G>A | p.(Glu101Lys) | c.1706-10G>A | p.(=) |
| c.313+1G>A | p.(=) | c.1796T>C | p.(Leu599Ser) |
| c.1706-1G>A | p.(=) | c.1436T>C | p.(Leu479Pro) |
| c.1706-1G>A | p.(Cys677Arg) | c.1474G>A | p.(Asp492Asn) |
| c.2029T>C | p.(Pro685Leu) | c.501C>A | p.(Cys167*) |
| c.2054C>T | p.(Trp483Arg) | c.662A>G | p.(Asp221Giy) |
| c.1447T>C | p.(Gly478Arg) | c.682G>T | p.(Glu228*) |
| c.1447T>C | p.(Asp72Thrfs*134) | c.1150C>T | p.(Gln384*) |
| c.214delG | p.(Trp87Gly) | c.938G>A | p.(Cys313Tyr) |
| c.259T>G | p.(Arg633Cys) | c.136T>G | p.(Cys46Gly) |
| c.1897C>T | p.(Asp227Glu) | c.2042G>C | p.(Cys681Ser) |
| c.681C>G | p.(Asn688Glnfs*29) | c.1618G>A | p.(Ala540Thr) |
| c.1285G>A | p.(Val429Met) | c.680_681delAC | p.(Asp227Glyfs*12) |
| KRAS, BRAF, PIK3CA ARRAY | |||
| KRAS Gene Targets | |||
| G12A | G12R | G12D | G12C |
| G12S | G12V | G13D | G13C |
| G13R | Q61K | Q61L | Q61R |
| Q61H(1) | Q61H(2) | A146T | A146P |
| BRAF Gene Targets | |||
| V600EG12R | |||
| PIK3CA Gene Targets | |||
| E542K | E545K | H1047R | |
| RESPIRATORY MULTIPLEX ARRAY II | |||
| Viral | |||
| Influenza A | Influenza B | Adenovirus A/B/C/D/E | Bocavirus 1/2/3 |
| Coronavirus 229E/NL63 | Coronavirus OC43/HKUI | Enterovirus A/B/C | Metapneumovirus |
| Parainfluenza virus 1 | Parainfluenza virus 2 | Parainfluenza virus 3 | Parainfluenza virus 4 |
| Respiratory syncytial virus A/B | Rhinovirus A/B/C | ||
| Bacterial | |||
| Legionella pneumophila | Bordetella pertussis | Chlamydophila pneumoniae | Haemophilus influenzae |
| Moraxella catarrhalis | Mycoplasma pneumoniae | Streptococcus pneumoniae |
| SEXUALLY TRANSMITTED INFECTION (STI) ARRAY | |||
| Pathogens | |||
| Chlamydia trachomatis (CT) | Neisseria gonorrhoea (NG) | Trichomonas vanginalis (TV) | Mycoplasma genitalium (MG) |
| Treponema pallidum (Syphilis) (TP) | Herpes simplex virus 1 (HSV-1) | Herpes simplex virus 2 (HSV-2) | Haemophilus ducreyi (HD) |
| Mycoplasma hominis (MH) | Ureaplasma urealyticum (UU) | ||
| URINARY TRACT INFECTION (UTI) ARRAY | |||
| Bacterial | |||
| Acinetobacter baumannii | Citrobacter freundii | Citrobacter koseri | Klebsiella aerogenes |
| Enterobacter cloacae | Enterococcus faecalis | Enterococcus faecium | Escherichia coli |
| Klebsiella oxytoca | Klebsiella pneumoniae | Morganella morganii | Proteus spp. |
| Pseudomonas aeruginosa | Providencia rettgeri | Providencia stuartii | Serratia marcescens |
| Staphylococcus aureus | Staphylococcus epidermidis | Staphylococcus saprophyticus | Streptococcus agalactiae (GBS) |
| Fungal | |||
| Candida albicans | |||
| Antibiotic Resistance Markers | |||
| mecA (incl MRSA) | Trimethoprim Resistance 1 | Trimethoprim Resistance 2 | Trimethoprim Resistance 3 |
| S mecA (incl MRSA) | Trimethoprim Resistance 4 | Trimethoprim Resistance 5 | Van A (Vancomycin Resistance A) |
| Van B (Vancomycin Resistance B) |
Related Services
Randox supports calls from Oxford University for more accurate diagnosis of diabetes following report warning
Calls for more accurate diagnosis of people at risk of developing Type-2 diabetes have been supported by Randox, following a warning raised by an Oxford University study which looked into efforts to tackle the worsening epidemic of the condition.
The study, which was published in the British Medical Journal, examined results from the NHS’s programme which involves a screening test for pre-diabetes. The authors determined that the UK’s National Diabetes Prevention Programme is unlikely to have much impact because the blood tests used were inaccurate at detecting pre-diabetes, though these are currently the only ones available to doctors and patients. The study argues that if the screening is inaccurate then people will either be falsely reassured or receive incorrect diagnoses, which will not help the worldwide challenge to reduce people at risk of developing diabetes that continues to increase across the world.
It is estimated that Type-2 diabetes causes 22,000 early deaths every year in England alone. Across the UK over 3m people currently have the condition though experts say this will increase to 5m by 2025.
With current treatment taking up almost 9% of the annual NHS budget – roughly £8.8bn a year – the implications for future healthcare budgets are clear if this dangerous trend persists.
Global reagents Manger Susan Hammond said,
“Although we wholly back the NHS’s belief that positive lifestyle changes make crucial differences in people’s health and lives, we also believe that unless earlier and more accurate diagnostic screening is employed on a twin-track of treatment, this epidemic will continue to worsen. We welcome that this study highlights the fact that clinician’s s are currently limited in what they can use to tackle the threat posed by diabetes. There are emerging biomarkers they could be given access to, such as Adiponectin and determining a person’s risk of Metabolic Syndrome.”
Assessing Adiponectin levels allows doctors to calculate the amount of visceral fat stored around a patient’s organs. This deep fat, which is not visible to the naked eye, is linked to health problems including Type-2 diabetes. High levels of adiponectin equate to low levels of visceral fat which can be combated by improving your diet, exercise habits and even stress levels. Given that 70% of Type-2 diabetes can be prevented by lifestyle changes, there is strong correlation that by detecting low levels of Adiponectin and taking corrective and preventive action, it could results in a decrease in the numbers of people who develop the life altering condition.
In addition to a test for the Adiponectin biomarker, Randox Biosciences have created a Metabolic Syndrome Array that measures 12 markers associated with metabolic syndrome and cardiovascular disease. Metabolic Syndrome is a is a group of cardiovascular risk factors that affects over 20% of adults and results in a person being three times more likely to have a stroke or heart attack, and five times more likely to develop diabetes.
Mrs Hammond concluded,
“We would ultimately like to see all medical professionals who are at the forefront of patient care armed with the most accurate diagnostic tools available. Updating traditional practice may not be easy but we believe it is imperative to do so, if we are to effectively challenge this global epidemic.”
Randox remains focused on providing early diagnoses and preventing illnesses by providing innovative diagnostics tests that will continue to revolutionise the healthcare landscape.
To find out more about our tests for metabolic arrays click here and Adiponectin click here.





